Liquid biopsy analysis detects cell-free DNA (cfDNA) released into the bloodstream by necrotic and apoptotic cells and thus offers an optimal alternative when tumor tissue is unavailable. However, only a fraction of circulating DNA originates from the tumor itself. Therefore, highly sensitive methods are required to detect these minimal ctDNA concentrations.
We at CeGaT established our CancerDetect® panel using duplex UMI-based technology, which detects sequence variants in actionable, most prevalent hotspots. Moreover, due to non-invasive and repeatable sampling, CancerDetect® represents a great approach to monitoring. By very sensitive detection of tumor-specific biomarkers, the analysis of ctDNA can be used as a surrogate marker for treatment response during medical follow-up.
Are you insured in Germany? Our colleagues at the Zentrum für Humangenetik Tübingen will gladly support you!
Ideally Suited for Monitoring and Follow-up
Our Promise to You
Service Details
- Liquid biopsy enables the detection of variants with potential therapeutic relevance in patients where the tumor is inaccessible, allowing to gain information about the tumor and to address treatment. — Learn more
- Highly sensitive and accurate detection of actionable, most prevalent driver mutations in thirty-six genes with very low allele frequencies by using duplex UMI-based technology (NAF ≥ 0.25%)
- High coverage: 50,000–100,000x raw coverage
- Simple, non-invasive, and repeatable sampling provides best conditions for longitudinal follow-up testing and disease monitoring.
- By very sensitive detection of tumor-specific biomarkers, the analysis of ctDNA can be used to track tumor dynamics in real time and intervene or adjust treatment if necessary (e. g., at acquired drug resistance).
- A list of all eligible drugs, with EMA and/or FDA approval, for which corresponding biomarkers could be detected in the tumor — learn more
We will report all relevant findings in a medical report. This includes a list of all identified clinically relevant variants. These variants are clearly annotated with gene name, functional category of the mutation, transcript-ID, allele frequency, and effect on protein function.
Each single medical report is prepared and discussed by an interdisciplinary team of scientists and physicians to guarantee highest quality.
Sample Report
Our Standard Sample Requirements
Liquid Biopsy
- 3x 10 ml cfDNA tubes for liquid biopsy
Notes on Sample Shipment
Here you can find more information on how to ship your sample safely.
This Is What Makes Our CancerDetect® Service Special
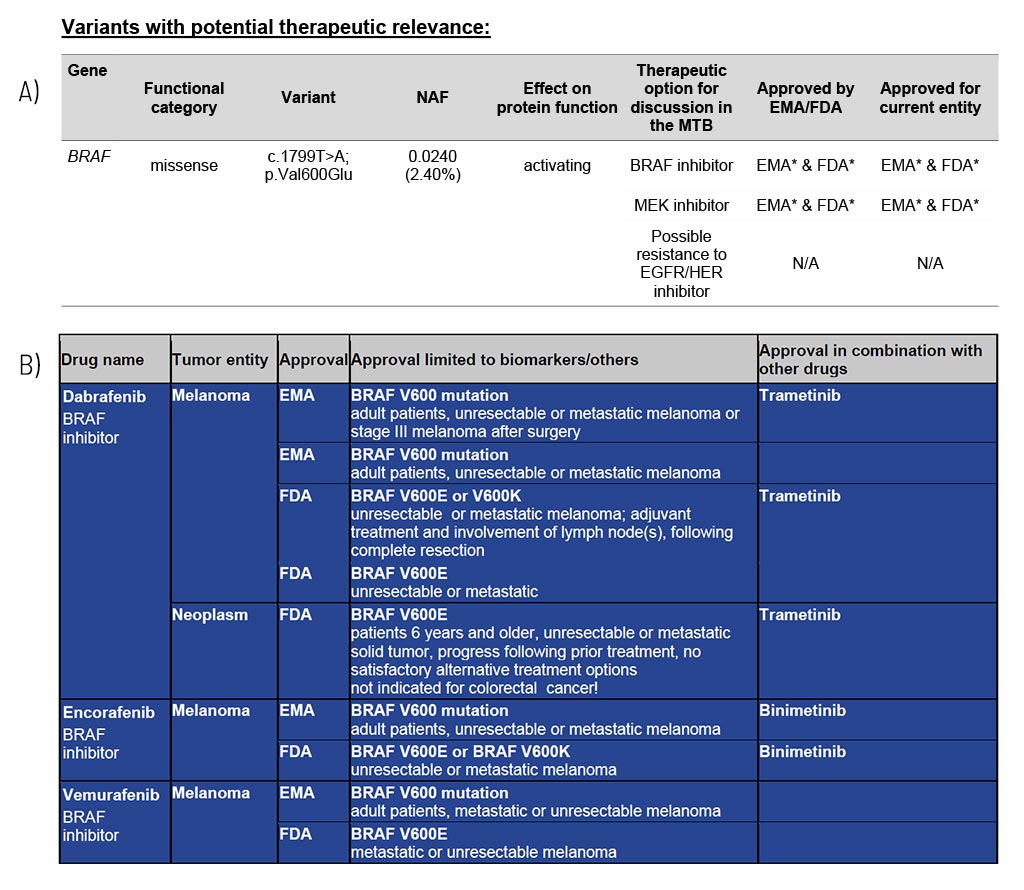
Variants with Potential Therapeutic Relevance
Guidance on potentially effective drugs
For each gene, the somatic change is depicted in detail, and the resulting therapeutic options are stated, including the EMA/FDA approval (A). These options are the basis for discussion in a molecular tumor board (MTB).
At the end of the medical report, in the appendix/supplement, we provide an extensive list of possible therapeutic strategies for each identified somatic changes (B). This list includes drug classes and names as well as their approval (FDA/EMA) and limiting conditions.
Sample Report: Exemplary for the BRAF variant detected and the resulting therapeutic options in a patient with melanoma. Top panel (A): An excerpt from Table 1 of the findings, listing variants with therapeutic relevance. Lower part (B): An excerpt of the drug listing. In addition to the drugs shown, other drugs are also described.
CancerDetect® Monitoring Applications
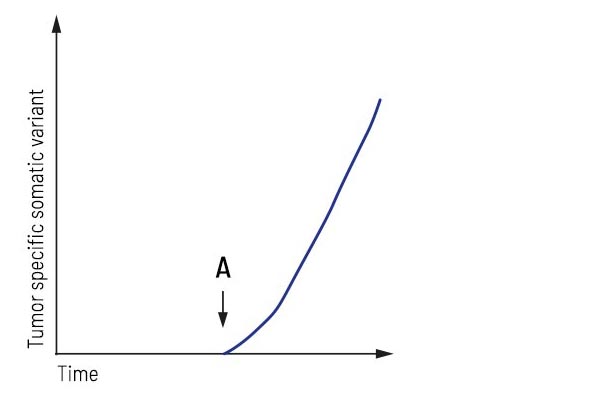
Application “Relapse detection”:
After surgery/treatment, the patient was considered tumor-free. Regular liquid biopsy testing revealed tumor progression, showing that an increase in a tumor-specific variant accompanied the recurrence of the tumor. This biomarker’s highly sensitive detection may detect the tumor’s relapse earlier than conventional imaging techniques.
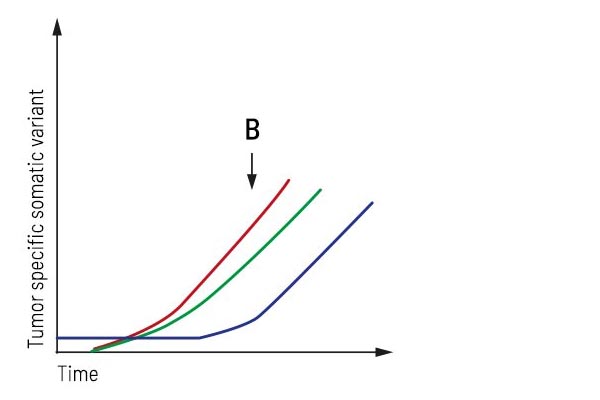
Application “Monitoring during treatment”:
After treatment, the patient underwent strong regression showing stable disease. Longitudinal monitoring provides an up-to-date molecular profile of the tumor and detects emerging treatment resistance in time. Here, two additional subclones occur besides the primary tumor mutation, forcing the treatment to be adjusted appropriately.
CancerDetect® Sample Case
Patient and indication:
- 42 years old, female, metastatic non-small cell lung cancer (NSCLC) with an EGFR L858R mutation in the primarius
- relapse after initial response to afatinib treatment
- recurrent tumor inoperable, no tumor biopsy possible
Primary finding:
The result of an analysis of cell-free DNA using a standard lung cancer panel remained negative.
CancerDetect® finding:
Our CancerDetect® analysis revealed a 2% tumor content in the liquid biopsy and detected the known EGFR L858R mutation, as well as an additional EGFR T790M resistance mutation. The EGFR T790M mutation represents one of the most common resistance mechanisms to tyrosine kinase inhibitors (TKIs) and typically occurs in NSCLC patients after first-line TKI treatment. In this patient, treatment was adjusted with the third-generation EGFR inhibitor osimertinib.
Gene Directory
All relevant variants in a named exon are analyzed. Exon numbers refer to coding exons (CDS) of the respective gene. The diagnostic is not limited to the listed example hotspot mutations. Exons not named and all variants within are not part of the analysis.
Gene | NM_Nr. | Enriched region (incl. example hotspot (HS)-variants) |
AKT1 | NM_005163 | Exon 2 (HS E17) |
ALK | NM_004304 | Exons 21-25 (incl. HS F1174) |
ARAF | NM_001654 | Exon 6 (HS S214) |
BRAF | NM_004333 | Exons 11 and 15 (incl. HS V600) |
CTNNB1 | NM_001904 | Exon 2 (incl. HS S37, S45) |
EGFR | NM_005228 | Exons 18-21 (incl. HS E746_A750del, T790, L858) |
ERBB2 | NM_004448 | Exon 8, 19-21 (incl. HS V842) |
ERBB3 | NM_001982 | Exons 3, 6-9, 23 (incl. HS V104, E928) |
ERBB4 | NM_005235 | Exon 12 (incl. HS E452) |
ESR1 | NM_000125 | Exons 4-8 (incl. HS K303, Y537, D538) |
FGFR2 | NM_000141 | Exons 6, 8, 11 (incl. HS S252, N549) |
FGFR3 | NM_000142 | Exon 12 (HS V555) |
GNA11 | NM_002067 | Exon 5 (HS Q209) |
GNAQ | NM_002072 | Exon 5 (HS Q209) |
GNAS | NM_000516 | Exon 8 (HS 201) and Exon 9 (HS Q227) |
H3-3A | NM_002107 | Exon 1 (HS K27 and G34) |
H3-3B | NM_005324 | Exon 1 (HS K37) |
HRAS | NM_005343 | Exons 1-3 (incl. HS G12, Q61) |
IDH1 | NM_005896 | Exon 2 (HS R132) |
Gene | NM_Nr. | Enriched region (incl. example hotspot (HS)-variants) |
IDH2 | NM_002168 | Exon 4 (HS R140, R172) |
JAK2 | NM_004972 | Exon 12 (HS V617) |
KIT | NM_000222 | Exons 9, 11, 13, 14, 17, 18 (incl. HS W557_K558del, D816) |
KRAS | NM_004985 | Exons 1-3 (inkl. HS G12, Q61) |
MAP2K1 | NM_002755 | Exon 3 (HS P124) |
MET | NM_001127500 | Exon 18 (incl. HS Y1248, Y1253) |
MYCN | NM_005378 | Exon 1 (HS P44) |
NRAS | NM_002524 | Exons 1-3 (inkl. HS G12, Q61) |
PDGFRA | NM_006206 | Exons 4, 9, 11, 13, 17 (incl. HS D842) |
PIK3CA | NM_006218 | Exons 4, 7, 9, 13, 20 (incl. HS E542, E545, H1047) |
PTEN | NM_000314 | Exons 5-7 (incl. R130, R233) |
RAC1 | NM_018890 | Exon 2 (HS P29) |
RAF1 | NM_002880 | Exon 6 (incl. HS S257, S259) |
RET | NM_020975 | Exon 10, 11, 13-16 (incl. HS C634) |
STAT5B | NM_012448 | Exon 15 (HS N642) |
TERT | NM_198253 | Promotor HS c.-124 (C228), c.-146 (C250) |
TP53 | NM_000546 | Whole coding region |
Further Information
Webinar: Discover the Power of Modern Tumor Diagnostics
Liquid biopsy in genetic tumor diagnostics – CancerDetect®
Contact Us
Do you have a question, or are you interested in our service?
Diagnostic Support
We will assist you in selecting the diagnostic strategy – for each patient.