The blood-based analysis of circulating tumor cells or tumor-derived nucleic acids is referred to as liquid biopsy. The main target of this analysis is cell-free DNA (cfDNA), which is released into the bloodstream by necrotic and apoptotic cells. Elevated levels of cfDNA are found in patients with cancer and other types of diseases. Since only a small fraction of the circulating DNA is derived from the tumor (circulating tumor DNA = ctDNA), highly sensitive detection methods are required.
Liquid biopsy applications are manifold, with several striking advantages over conventional tissue analysis. Most importantly, the analysis of liquid biopsies is based on a simple blood draw and, thus, easily repeatable.
Application areas of liquid biopsies:
- monitoring of tumor disease
- monitoring of treatment response
- patient stratification and treatment selection
- detection of minimal residual disease
- early detection and profiling of resistance to therapy
We offer different Liquid Biopsy products to get reliable and accurate insights into cell-free DNA.
CeGaT Is the Best Partner for Sequencing Your Project
Our Commitment to You
Fast Processing
Turnaround time
≤ 15 business days
High Quality
Highest accuracy for all processes
Secure Delivery
Secure provision of sequenced data via in-house servers
Safe Storage
Safe storage of samples and data after project completion
Our Service
We provide a comprehensive and first-class project support – from selecting the appropriate product to evaluating the data. Each project is supervised by a committed scientist. You will have a contact person throughout the whole project.
Our service includes:
- detailed project consulting
- product selection tailored to your project
- detailed bioinformatic evaluation of your data
- detailed project report with information about sample quality, sequencing parameters, bioinformatic analysis, and results
Benefit from our dedicated support and accredited workflows.
Explore Our Product Portfolio for Liquid Biopsies
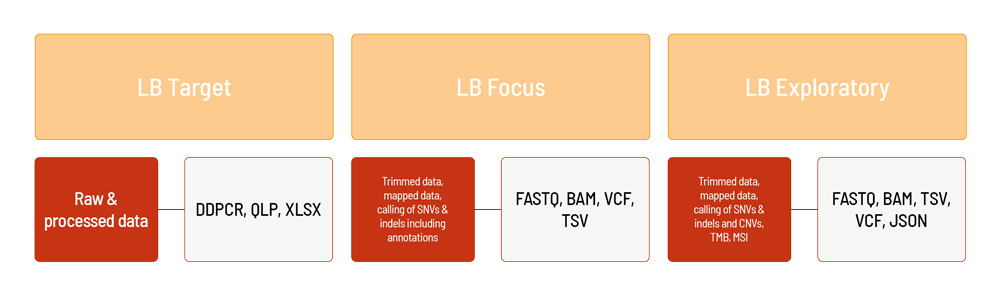
We offer different Liquid Biopsy products to address a variety of research questions. By selecting LB Target, you choose a digital droplet PCR approach. This approach especially suites the monitoring of single variants. LB Focus targets selected cancer-associated genes. This next-generation sequencing (NGS)-based approach uses a gene panel designed by CeGaT. When choosing LB Exploratory, you will receive comprehensive genomic profiling of cfDNA using the TSO500 ctDNA assay from Illumina. For LB Flex, either the whole exome or a pre-defined subset of genes can be targeted. Would you like to have bioinformatic analyses performed on your data in addition to the included deliverables? Each of our products can be supplemented with further services. We are happy to advise you.
Would you like to have bioinformatic analyses performed on your data in addition to the included deliverables? Each of our products can be supplemented with further services. We are happy to advise you.
LB Target | LB Focus | LB Exploratory | LB Flex |
Species | Species | Species | Species |
Starting material | Starting material | Starting material | Starting material |
Target | Target | Target | Target |
Technology | Technology | Technology | Technology |
Bioinformatic pipeline | Bioinformatic pipeline | Bioinformatic pipeline | Bioinformatic pipeline |
Duplex UMIs | Duplex UMIs | Duplex UMIs | Duplex UMIs |
Variant type | Variant type | Variant type | Variant type |
Included deliverables | Included deliverables | Included deliverables | Included deliverables |
Bioinformatics
For LB Target, droplet digital PCR results are available as raw data (DDPCR, QLP files) and processed XLSX files (e.g., copies/µl, droplet counts). In addition to the data, a project report is generated (PDF format).
For LB Focus, the raw sequencing data (FASTQ format) are automatically processed based on CeGaT’s bioinformatic pipeline. The analysis for LB Focus includes the alignment of the demultiplexed and adapter trimmed sequencing data (BAM format), as well as calling and annotation of SNVs and indels (VCF and TSV format). In addition to the data, a project report is generated (PDF format).
For LB Exploratory, the raw sequencing data (FASTQ format) are automatically processed based on the TSO500 (Illumina) pipeline. The analysis for LB Exploratoryincludes the alignment of trimmed sequencing data (BAM format), calling and annotation of SNVs and indels, as well as fusions (VCF, TSV, and JSON format). Furthermore, CNVs (VCF format) and the MSI/TMB status (TSV format) are evaluated. In addition to the data, a project report is generated (PDF format).
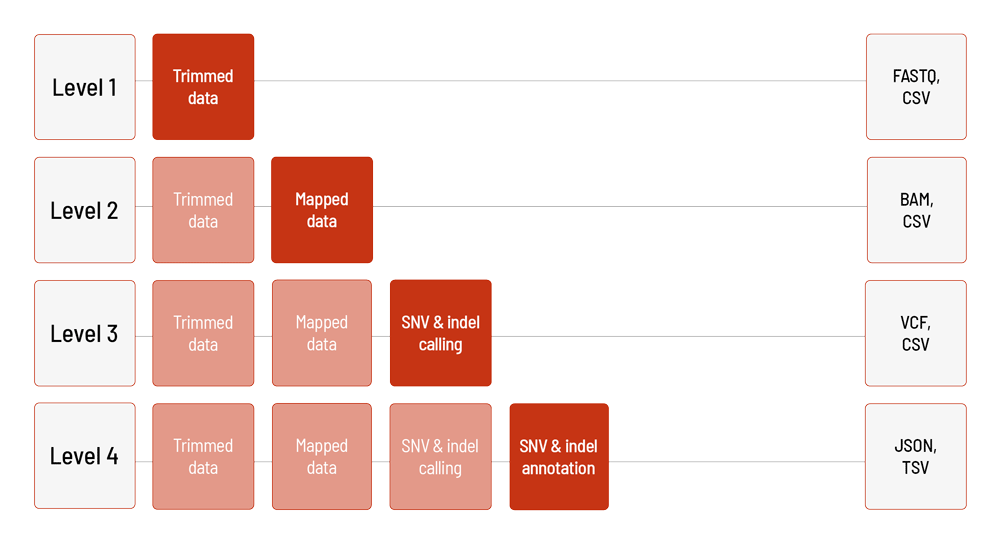
For LB Flex, the raw sequencing data are automatically processed based on the DRAGEN Bio-IT Platform. We offer different levels of bioinformatic analysis for LB Flex. The default level is Level 1. With increasing bioinformatic level, more data are delivered. All higher levels include the data from the lower levels. In addition to the data, and independent of the analysis level, a project report is generated.
Level 1:
- demultiplexing and adapter trimming of the sequencing data (FASTQ format)
- metrices (CSV format)
- MultiQC report (HTML format)
Level 2:
- mapping of the sequencing data (BAM format)
- metrices (CSV format)
Level 3:
- calling of single nucleotide variants (SNVs) and small insertions and deletions (indels) (VCF format)
- metrices (CSV format)
Level 4:
- annotation of the SNVs and indels (JSON and TSV format)
Technical Information
At CeGaT, the droplet digital PCR is performed using the QX200 Droplet Digital PCR System (Bio-Rad). The paired-end sequencing (2 x 100 bp) is performed using the Illumina sequencing platforms. If you require other sequencing parameters, please let us know! We can provide further solutions.
Gene Directory for LB Focus, LB Exploratory, & LB Flex (Panel)
Further Information about Liquid Biopsy
The analysis of cell free tumor DNA has several striking advantages over conventional tissue analysis:
- It always represents a current picture of the disease.
- It gives insight into the heterogeneity of the complete tumor and not just a small fraction of it.
- Most important, it is based on just a simple blood draw. This means that the test is patient friendly, non-invasive, and easily repeatable.
These advantages open a wide field of applications. We will give you two examples for possible applications.
For running a clinical trial, e.g., for a relatively new drug, one might want to know the possible effects, identify possible drug responders, understand the response mechanism behind it, identify potential biomarkers, track the evolutionary dynamics of the tumor, and uncover resistance mechanisms. We designed our Liquid Biopsy Exploratory product for such purposes. This product covers a large panel of more than 520 cancer associated genes as well as immuno-oncology biomarkers. Additionally, it makes use of so called UMIs and we sequence at a very high sequencing depth. Thus, the Liquid Biopsy Exploratory combines comprehensive genomic profiling from plasma on the one hand, with a high sensitivity on the other hand.
Another application area of liquid biopsy is the monitoring of a tumor disease and its treatment response. Liquid biopsies can be used, if the patient’s tumor and its driver mutations are already analyzed and known, and the patient shows stable disease or even complete response to the therapy, but has a risk for relapse. With liquid biopsies, a possible emergence of residual disease and recurrence can be detected as early as possible. For this purpose, our Liquid Biopsy Focus product is well suited. Instead of a large panel, it focuses on a relatively small number of selected genes to be analyzed. Nevertheless, maximum sensitivity is needed. Therefore, we make use of duplex UMIs combined with a high sequencing depth. That allows us to detect variants down to 0.25% allele frequency.
Those were just two examples, and we understand that each project and its demands are different. That is why we offer several services for liquid biopsy analysis, and we are happy to guide you in finding the optimal strategy for your project’s needs. We want to be your partner for translating liquid biopsy assays into clinical practice.
Video: Make Use of the Manifold Applications of Liquid Biopsies
Downloads
Contact Us
Do you have a question or are you interested in our service? Feel free to contact us. We will take care of your request as soon as possible.
Start Your Project with Us
We are happy to discuss sequencing options and to find a solution specifically tailored to your clinical study or research project.
When getting in contact, please specify sample information including starting material, number of samples, preferred library preparation option, preferred sequencing depth and required bioinformatic analysis level, if possible.