Small RNA sequencing focuses on short RNA molecules. These short RNA molecules play an important role in silencing and processes of post-transcriptional gene expression regulation. Sequencing of small RNAs allows the analysis of these molecules in more detail and, therefore, permits
- the profiling of all small RNAs and miRNAs in the transcriptome,
- the identification of novel small RNAs,
- the tissue-specific differential expression analysis of small RNAs, and
- the identification of novel biomarkers for cancer and other diseases.
Our Small RNA Sequencing products offer reliable and accurate insights into small RNAs.
CeGaT Is the Best Partner for Sequencing Your Project
Our Commitment to You
Fast Processing
Turnaround time
≤ 15 business days
High Quality
Highest accuracy for all processes
Secure Delivery
Secure provision of sequenced data via in-house servers
Safe Storage
Safe storage of samples and data after project completion
Our Service
We provide a comprehensive and first-class project support – from selecting the appropriate product to evaluating the data. Each project is supervised by a committed scientist. You will have a contact person throughout the whole project.
Our service includes:
- detailed project consulting
- product selection tailored to your project
- detailed bioinformatic evaluation of your data
- detailed project report with information about sample quality, sequencing parameters, bioinformatic analysis, and results
Benefit from our dedicated support and accredited workflows.
Explore Our Product Portfolio for Small RNA Sequencing
We offer different Small RNA Sequencing (SRS) products to address a variety of research questions. Would you like to have bioinformatic analyses performed on your data in addition to the included deliverables? Each of our products can be supplemented with further services. We are happy to advise you.
SRS Classic | SRS Flex |
Species | Species |
RNA quality & input | RNA quality & input |
Sequencing platform | Sequencing platform |
Output | Output |
Included deliverables | Included deliverables |
Bioinformatics
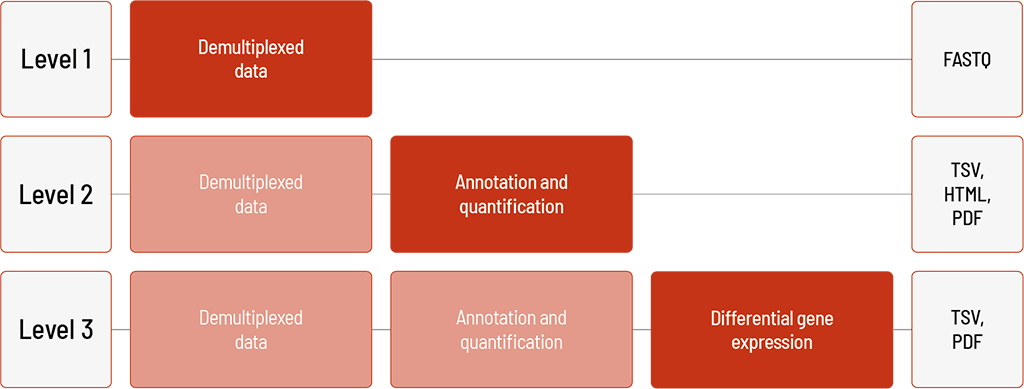
Raw sequencing data are automatically processed. We offer different levels of bioinformatic analysis. The default level is Level 1. With increasing bioinformatic level, more data are delivered. All higher levels include the data from the lower levels. In addition to the data, and independent of the analysis level, a project report is generated.
Level 1:
- demultiplexing of the sequencing data (FASTQ format)
Level 2:
- annotation with miRbase (TSV format)
- quantification of miRNAs (TSV format)
- experiment-wide comparison (HTML format)
- novel miRNA prediction (TSV and PDF format)
Level 3:
- normalization (TSV format)
- differential gene expression analysis (reads can be mapped to known miRNAs and pre-miRNAs) (TSV format)
- visualization (PDF format)
⚠ For the differential gene expression analysis (group comparison), a minimum of three replicates per group is required.
Technical Information
Small RNA Sequencing is performed using the Illumina sequencing platforms in single-read mode (1 x 72 bp) with at least ten million clusters per sample. If you require other sequencing parameters, please let us know! We can provide further solutions.
Further Information about Small RNA Sequencing
Small RNA sequencing, also known as small RNAseq or sRNAseq, uses high-throughput sequencing technologies to get information about small non-coding RNA molecules. Small RNAs have a length between twenty and 200 nucleotides and are usually non-coding. These small non-coding RNAs include small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), small conditional RNA (scRNA), piwi-interacting RNA (piRNA), microRNA (miRNA), YRNA (components of the Ro60 ribonucleoprotein particle), tRNA-derived small RNA (tsRNA), rRNA-derived small RNA (rsRNA), and small interfering RNA (siRNA). The function of these small RNAs range from RNA interference, also known as RNAi, over RNA processing and modification, to gene silencing, protein stability and transport, and epigenetic modifications.
Thus, studying small RNAs, has a wide range of applications. With small RNA sequencing, the differential expression of all small RNAs in a sample can be determined. This also includes the expression profile of miRNAs and other small RNAs. By analyzing those expression profiles using small RNA sequencing, the understanding of cell regulation and dysregulation under pathological conditions can be increased. Thus, small RNA sequencing can be used to identify biomarkers for disease diagnostics or disease classification, such as cancer. As small RNAs play a crucial role in regulations, regulatory networks can be identified to increase the knowledge about developmental mechanisms of
tissues and organisms. Additionally, with small RNA sequencing, novel small RNAs can be discovered. These findings can further be used for the prediction of small RNAs.
miRNAs can be of particular interest, especially in the context of diseases. MicroRNAs, abbreviated as miRNAs, are small non-coding RNA molecules of approximately twenty-two nucleotides. miRNAs can be found in animals, plants, and some viruses. Their function is determined in RNA silencing and post-transcriptional regulation of the gene expression. By binding with a complementary mRNA sequence, miRNAs silence the respective mRNA molecules. As miRNAs play a pivotal role in the normal functioning of eukaryotic cells, dysfunctional miRNAs can be associated with disease. Thus, small RNA sequencing can give insights into disease emergence and development, including cancer, heart diseases, kidney diseases, or diseases of the nervous system. With small RNA sequencing, the expression of the miRNAs can be quantified, targeting genes of miRNAs can be predicted, and the understanding of miRNAs in particular, and small RNAs in general, can be increased. Thus, small RNA sequencing has valuable potential in the areas of diagnostics and prognostics.
Downloads
Contact Us
Do you have a question or are you interested in our service? Feel free to contact us. We will take care of your request as soon as possible.
Start Your Project with Us
We are happy to discuss sequencing options and to find a solution specifically tailored to your clinical study or research project.
When getting in contact, please specify sample information including starting material, number of samples, preferred library preparation option, preferred sequencing depth and required bioinformatic analysis level, if possible.